A Numerical Study of the Interaction between 1D Carbyne Chains and ssDNA
Student: Zeina Salman
Degree: Ph.D., August 2017
Major Professors: Dr. Steve Tung, Dr. Arun Nair
Research Area(s):
Biological Sensors
Modeling & Simulation
Background/Relevance
- Biosensors function based on the electrical and mechanical properties associated with the interaction between the sensing elements and biomolecules.
- Current applications of biosensors to sequence DNA lack the required spatial resolutions.
Innovation
- The research involves using the thinnest possible nanowire, carbyne, as a sensing element to increase the spatial resolution at the single molecule level
- Carbyne has performed extreme mechanical performance and double the stiffness of graphene and diamond.
Approach
- First approach is using first principle simulation. Quantum ESEPRESSO and its extension wannier90, open source codes, are used to calculate the quantum conductance and density of states that are used to calculate current associated with the interaction between carbyne chain and ssDNA
- Second approach is using molecular dynamic simulation (MD). Material Studio, available in lab, is used to calculate the deformation in chain caused by the presence of each ssDNA to estimate the changes in the electrical resistance of the chain.
Key Results
- First principle simulation using density functional theory (DFT) and non-equlibrium Green’s function (NEGF)
- Codes: Quantum ESPRESSO & wannier90
- Carbyne chain of 16 carbon atoms and carbyne chain of 16 carbon atoms attached to two graphene sheets on both sides
- ssDNA of one base at a time
- Molecular Dynamics simulation(MD)
- Code: Material Studio
- Carbyne chain of 16 carbon atoms
- ssDNA of one base at a time
- In two cases:
- Carbyne alone Analyzing the differences
- Carbyne with ssDNA
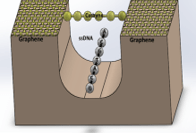
Conclusions
- Bases A G, C, and T of ssDNA have been included in two different structures that have carbyne. The quantum conductance, density of states, and current have been calculated and compared with the results when no amino acid included.
- Base A of ssDNA has been included in two different structures that have graphene. The transport properties have been calculated and compared with the results when no amino acid included. Also, the results have been compared with carbyne.
- Different locations and orientations of ssDNA have been considered to understand the types of interaction. Force model has been developed. The force’s trend has been compared with the current’s trend to validate the model. Both trends match nicely.
- MD simulation has been performed to estimate the changes in the electrical resistance in the chain and binding energy of ssDNA to the carbyne’s surface.
